Multiomic Analysis
We carry out the integration of different omics data
Home / Multiomic Analysis
Towards the future of biomedical research
We offer supervised integrative analysis of transcriptomic, proteomic and metabolomic data to help identify those biomarkers that most influence the clustering of the samples included in your experiment.
The bioinformatics analysis we perform at Dreamgenics includes:
- Personalized advice, both in the experimental design and in the interpretation of results.
- Reading and preprocessing of data, including column naming, filtering of irrelevant data and reordering according to specific criteria.
- Transformation and normalization of data to ensure comparability and prepare them for further analysis.
- Assessment of data quality through exploratory analysis, such as principal component analysis (PCA), to identify possible patterns or problems.
- Integration of data from different omics platforms to perform a joint analysis to obtain a global and coherent view of the data.
- Functional enrichment analysis to identify metabolic pathways and significant biological terms, providing biological context to the results observed, both in individual omics and in the integrated analysis.
- Identification and annotation of metabolites, facilitating interpretation and connection with public databases for their characterization.

Multiomic Analysis
Learn more about our multi-omics data integration analysis by downloading our brochure
Results in Genome One Reports
The results of all analyses result in interactive graphs that can be explored on our Genome One Reports platform and downloaded as PDFs for use in scientific publications.
- Correlation study between different omics datasets
- Individualized analysis of each omics: clustering and pathway enrichment
- Correlation Circle Plot, Multiblock Circle Plot, Relevance Networks, Loading Weights, Clustered Heatmap and Clustering Results charts.
- Enrichment of Gene Ontologies (WikiPathways, GO-MF, GO-CC and GO-BP) for each omics in all experimental groups.
- All tables and figures are easily exportable, ready to be used in scientific publications.
Preparation and Independent Analysis
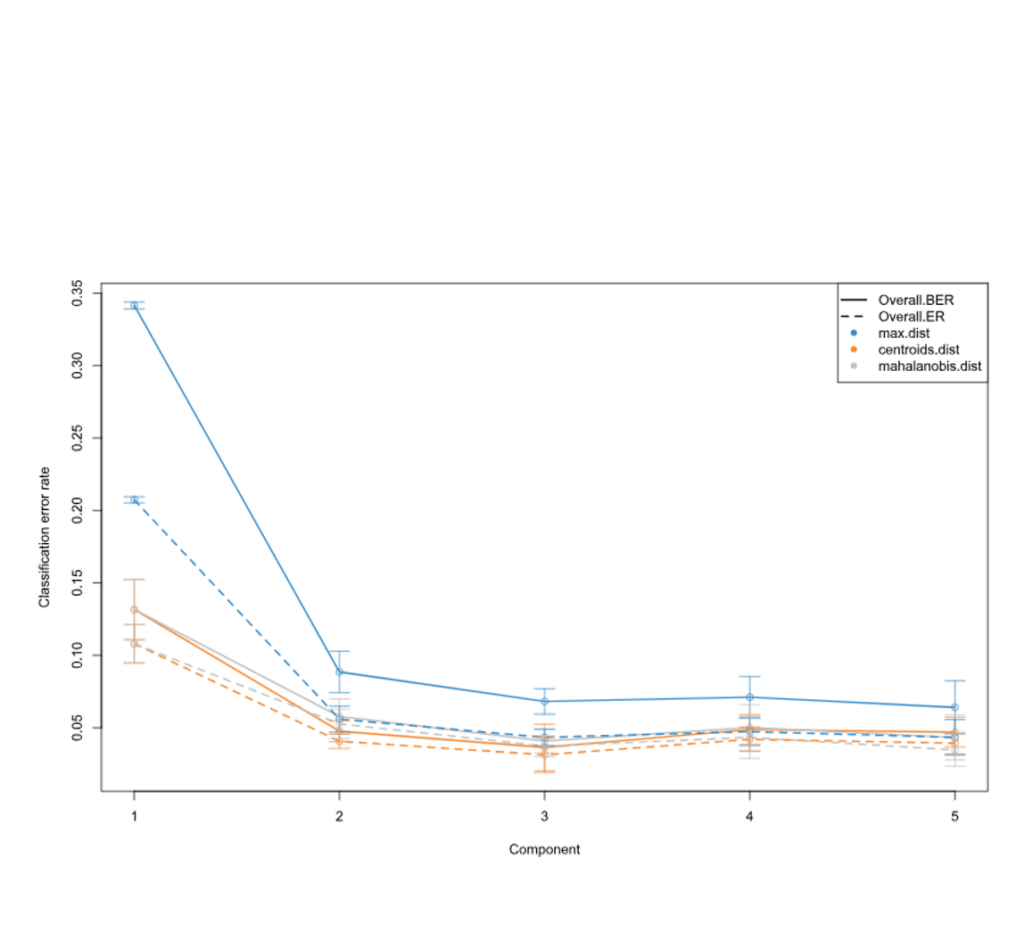
Error Rates
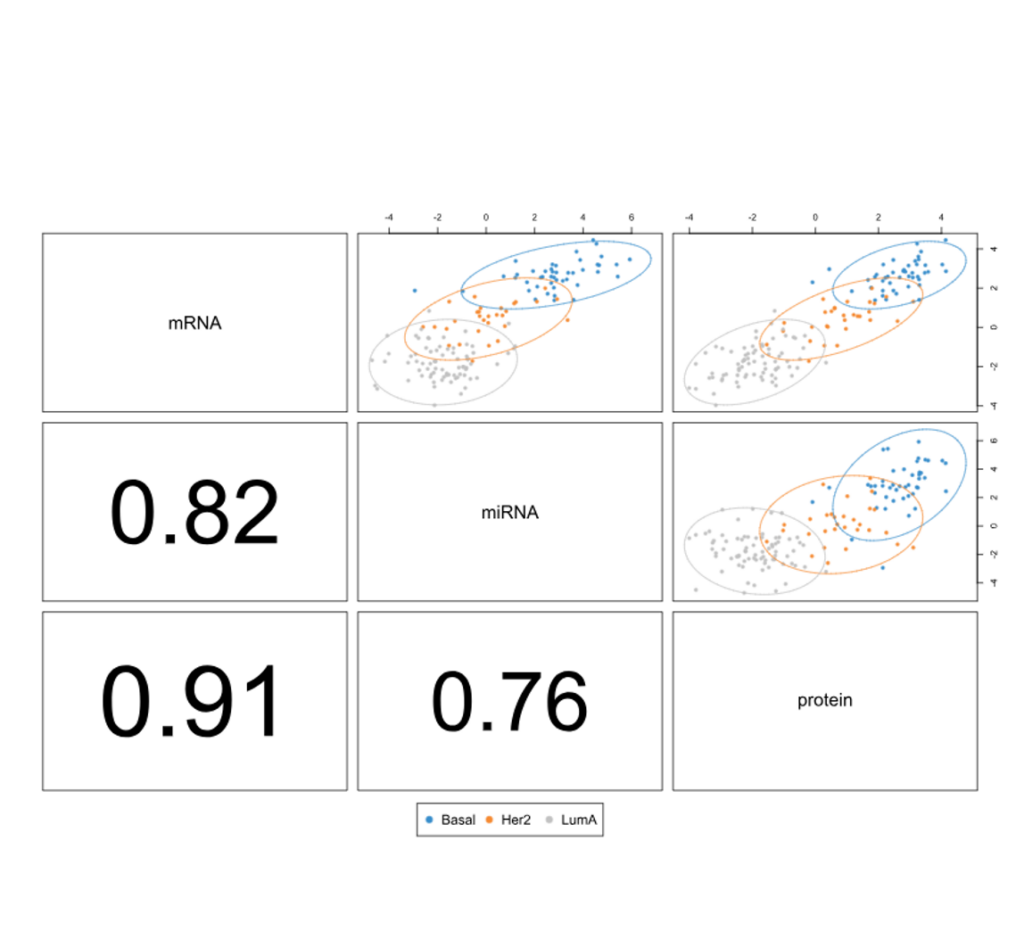
Correlation Coefficients
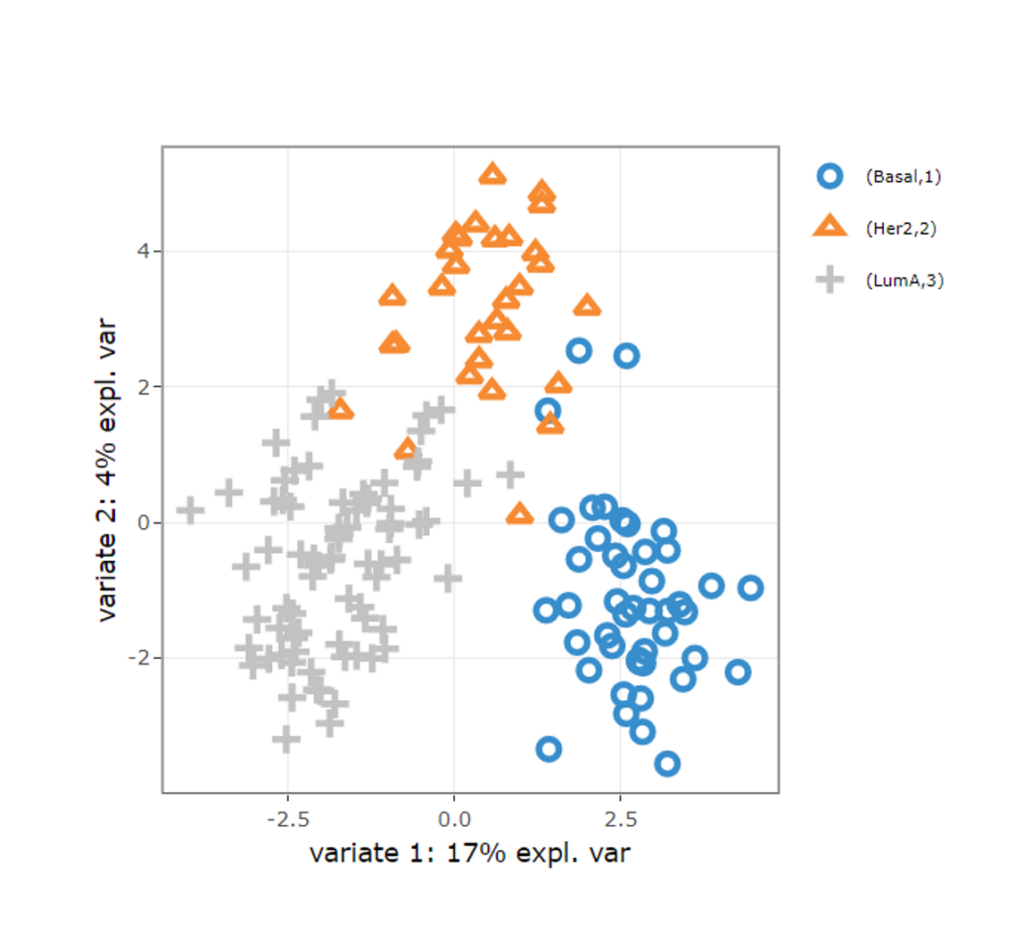
Individual Cluster
Integrated Analysis
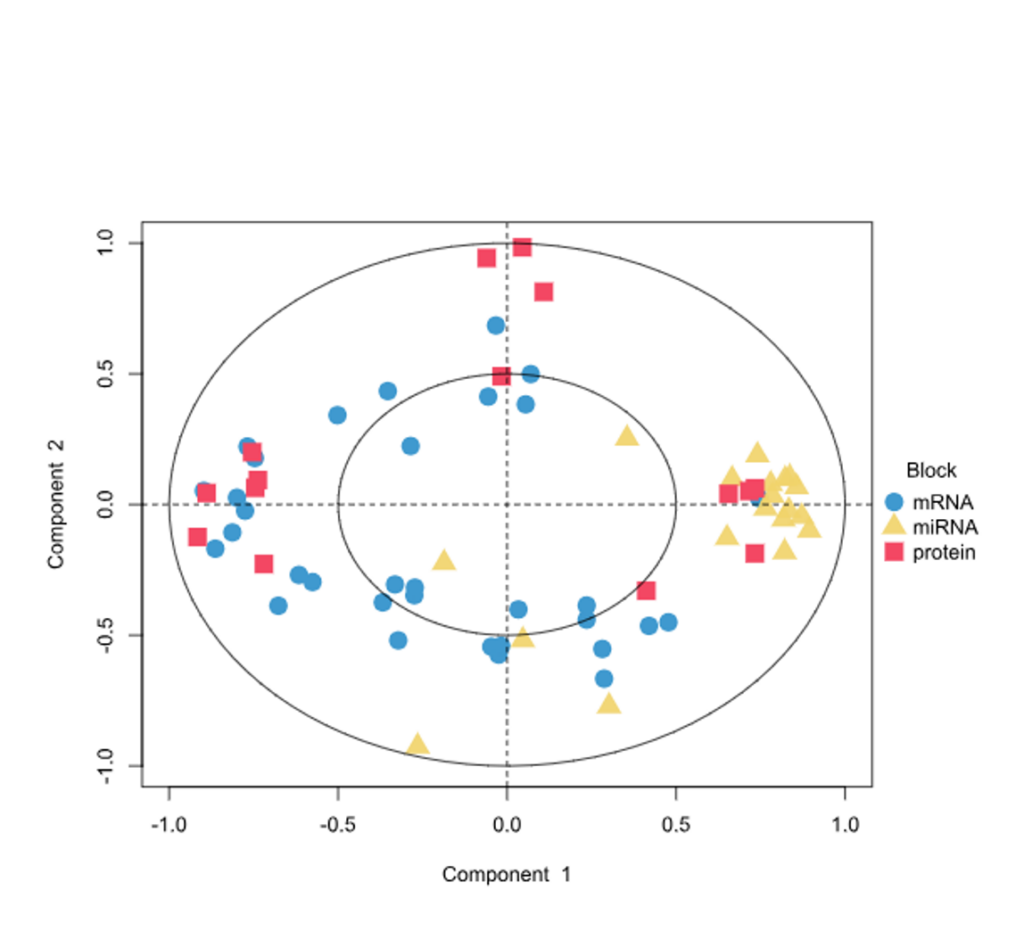
Correlation Circle Plot
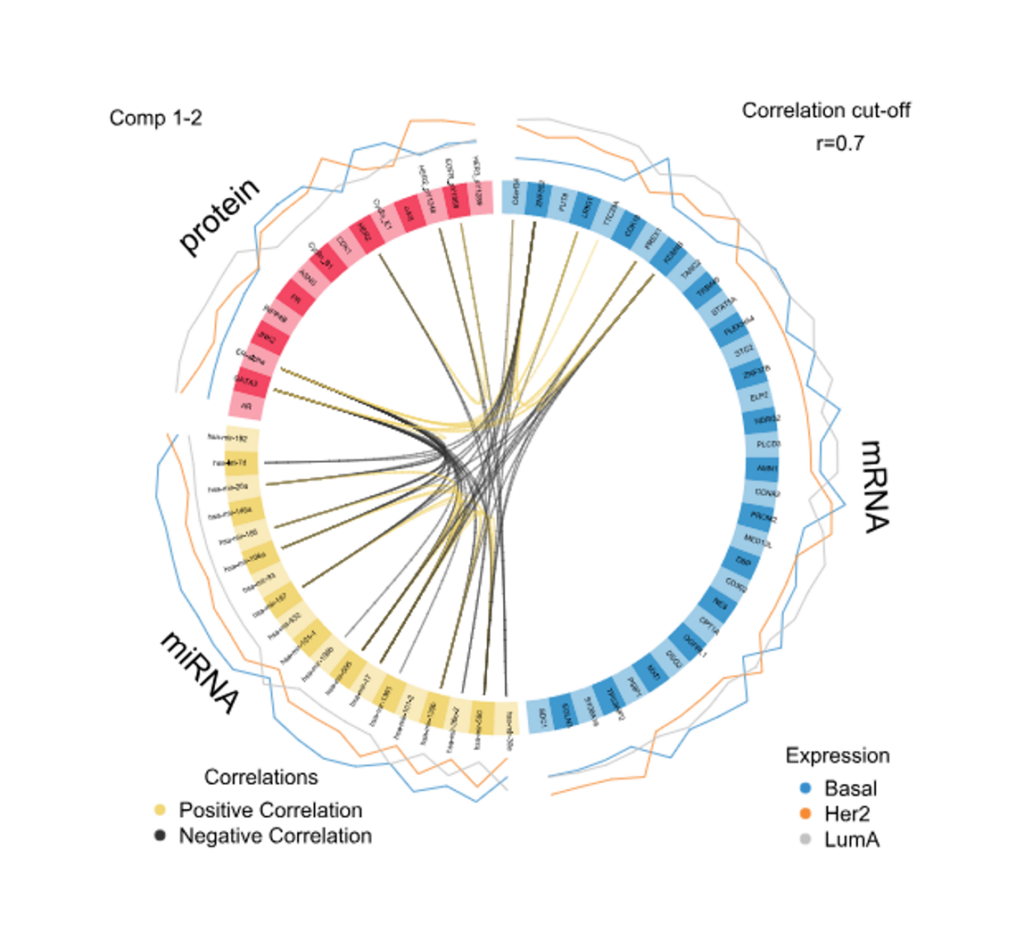
Plot Multiblock Circos
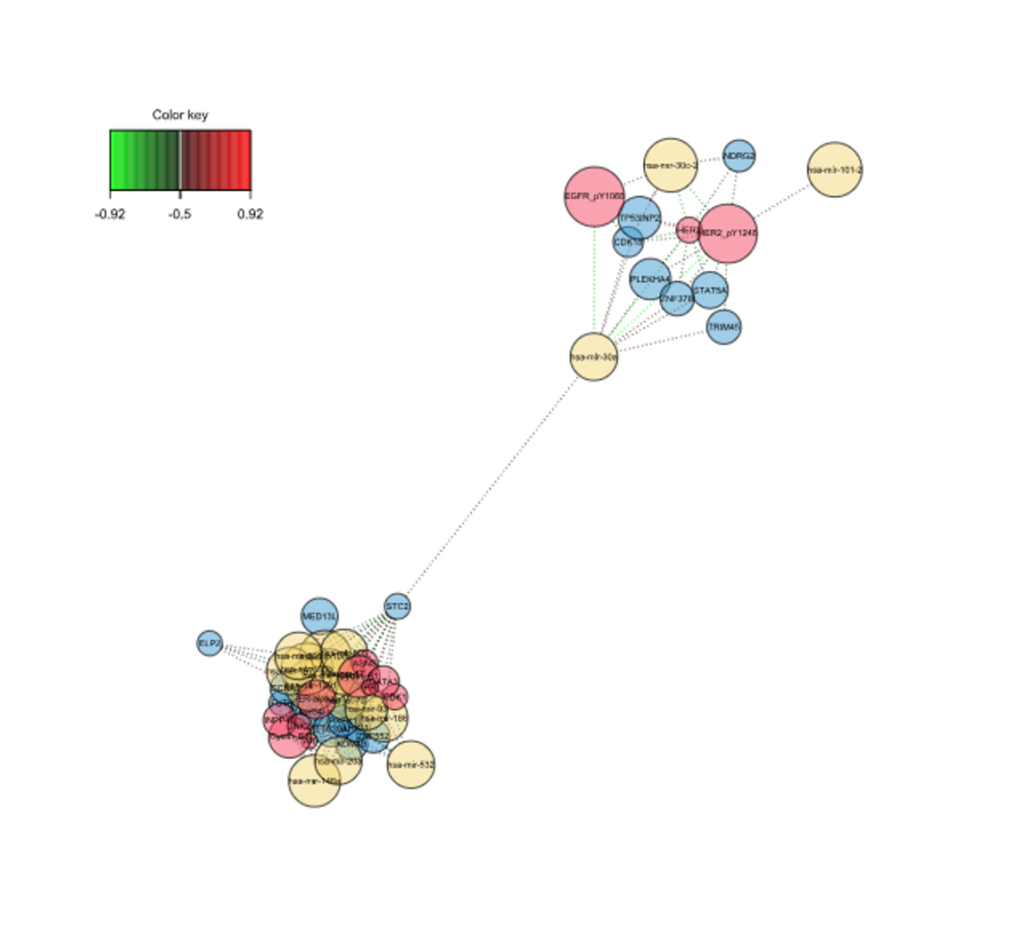
Relevance Networks
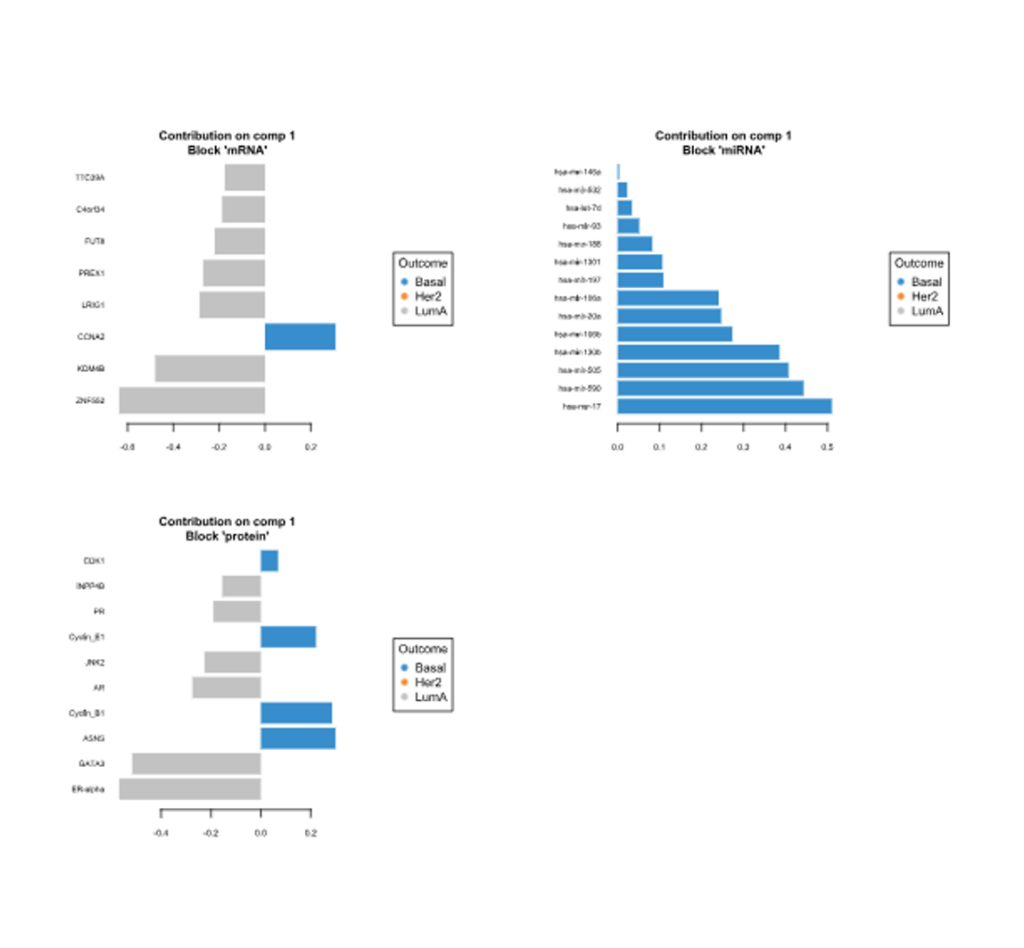
Loading Weights
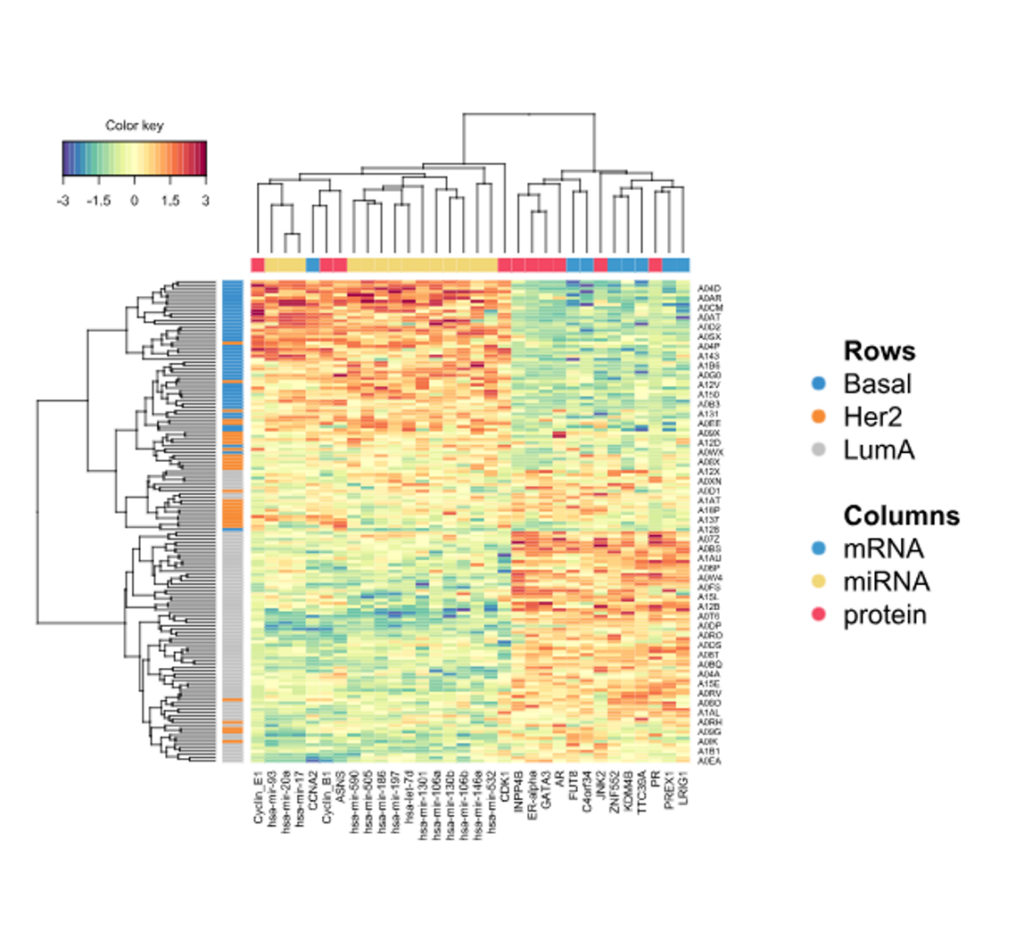
Clustered Heatmap
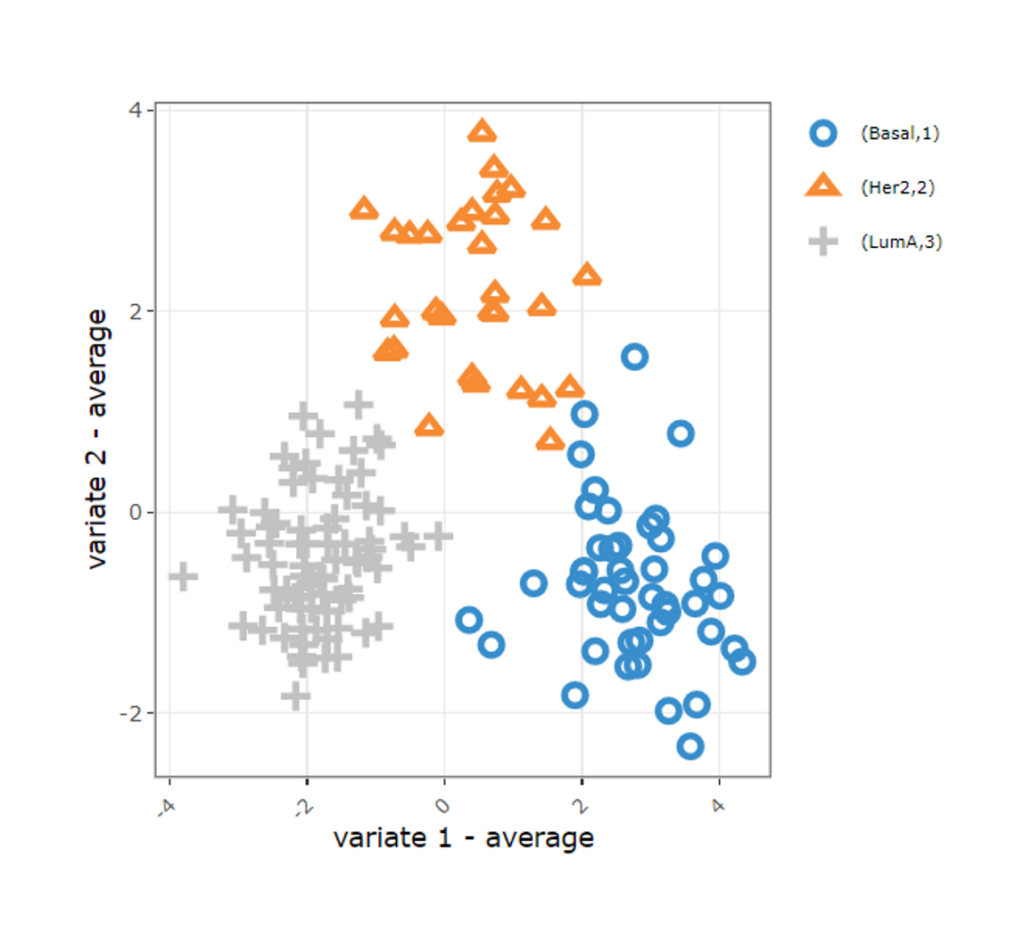
Clustering Results
"The possibility of integrating different datasets obtained in our laboratory through multi-omics analysis has opened up new avenues of research in the study of human diseases and for the identification of new biomarkers. Thanks to the integration of various omics data, including transcriptomics, proteomics and metabolomics, we obtain a better classification of patients and a more accurate characterization of the pathophysiology of the disease."

Dr. Héctor González Iglesias
Scientific researcher - CSIC

























