Single cell RNA-Seq
We offer bioinformatics analysis of NGS data for single cells.
Home / Single cell RNA-Seq
Study of gene expression at the cellular level
Single cell RNA-Seq analysis allows the transcriptome of each cell to be analyzed individually and can reveal specific biological processes and molecular mechanisms that might not be detected by RNA sequencing techniques at the population or tissue level.
The bioinformatics analysis we perform at Dreamgenics includes:
- Sequencing and mapping quality control.
- Alignment of reads against reference genome.
- Quantification of gene expression at the single cell level.
- Reduction of data dimensionality.
- Clustering or division of cells into different groups and subgroups.
- Differential gene expression analysis.
- Study of the evolutionary pattern or cell maturation (Velocity or Trajectory Analysis).
- Enrichment study, gene ontologies and pathways.

Bioinformatics Services
Learn more about our NGS data analysis services by downloading our brochure
Results in Genome One Reports
The results of all analyses result in interactive graphs that can be explored on our Genome One Reports platform and downloaded as PDFs for use in scientific publications.
- You will have access to the quality analysis of the sequences obtained.
- You will have access to the results of the Cell Ranger analysis.
- PCA plot, Cell Clustering, Trajectory Analysis, RNA Velocity, Cell Chat, etc. depending on the chosen package.
- Enrichment of Gene Ontologies paths and terms (WikiPathways, GO-MF, GO-CC and GO-BP)
- All tables and figures are easily exportable, ready to be used in scientific publications.
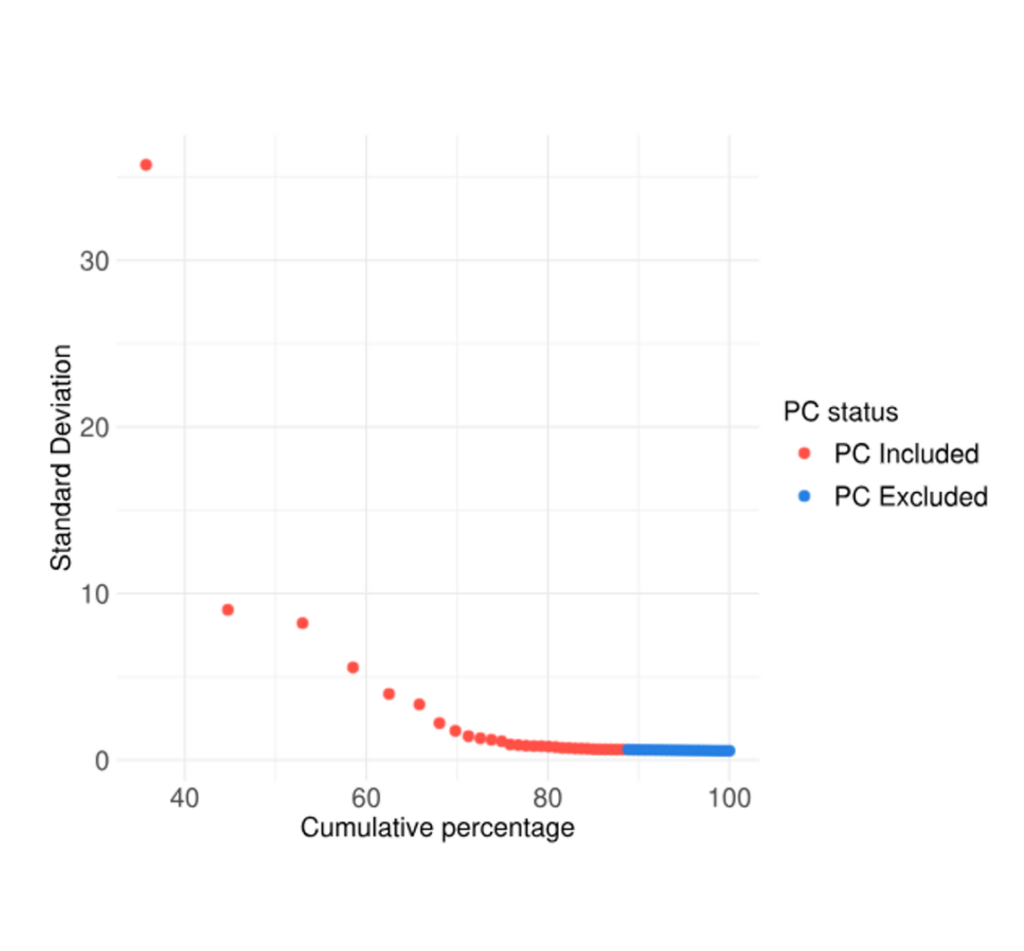
PCA Plot
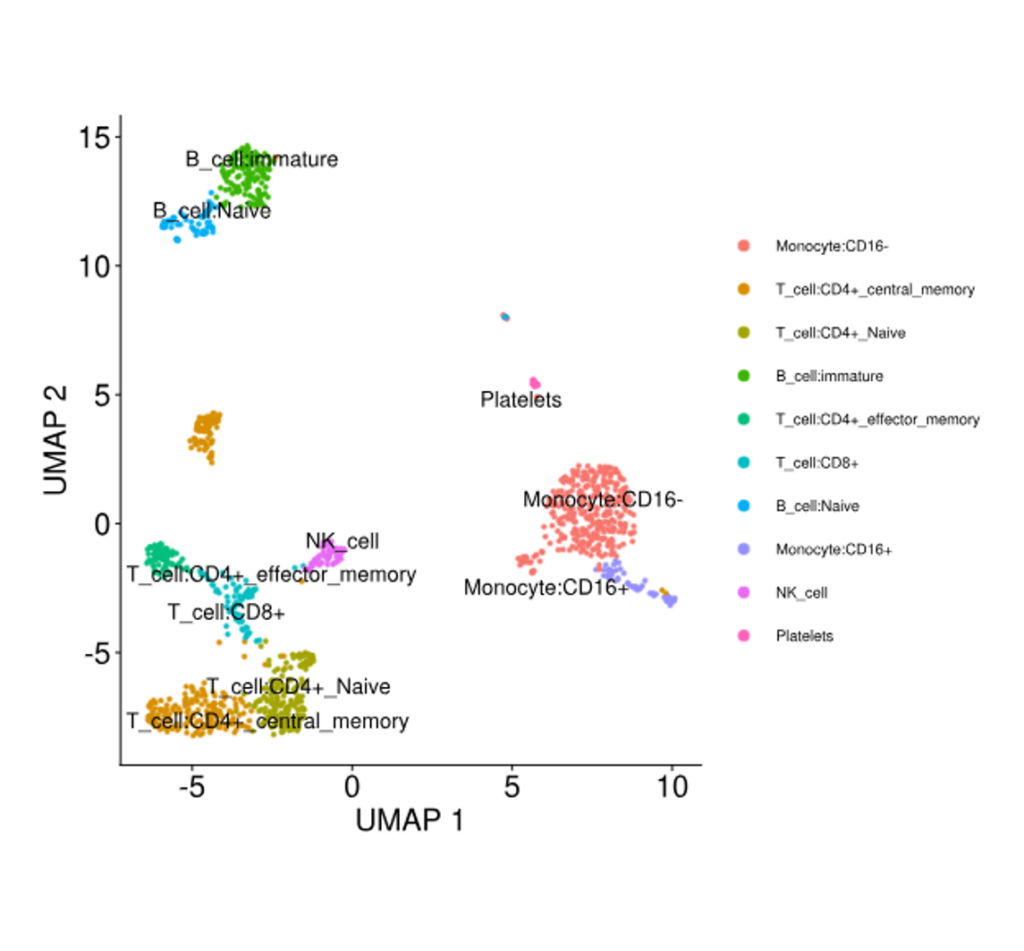
Cell Clustering
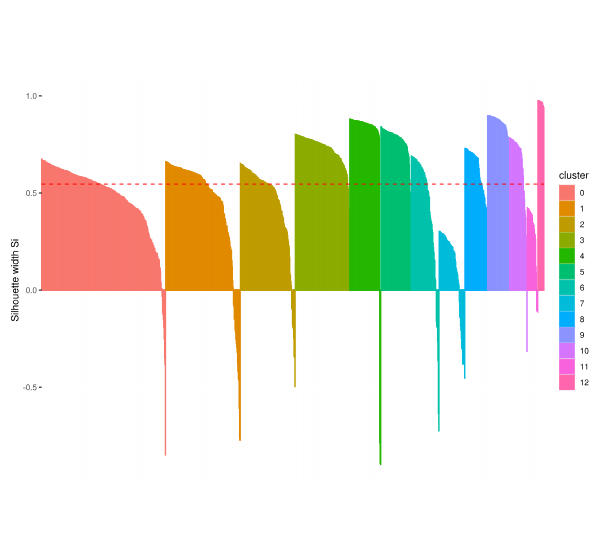
Cluster Evaluation
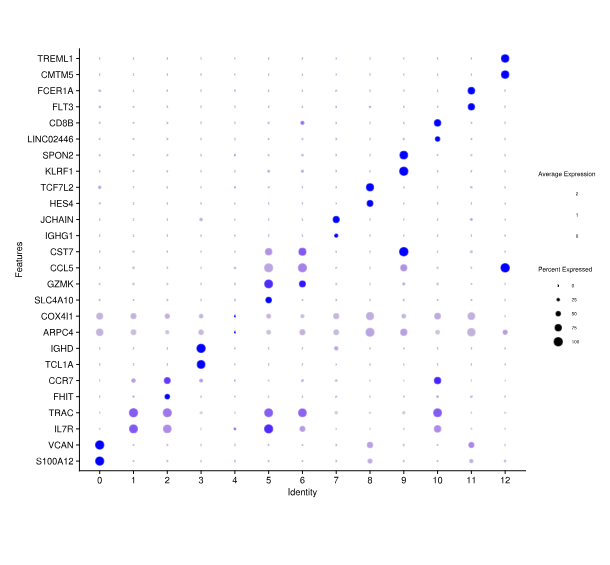
Differentially Expressed Genes
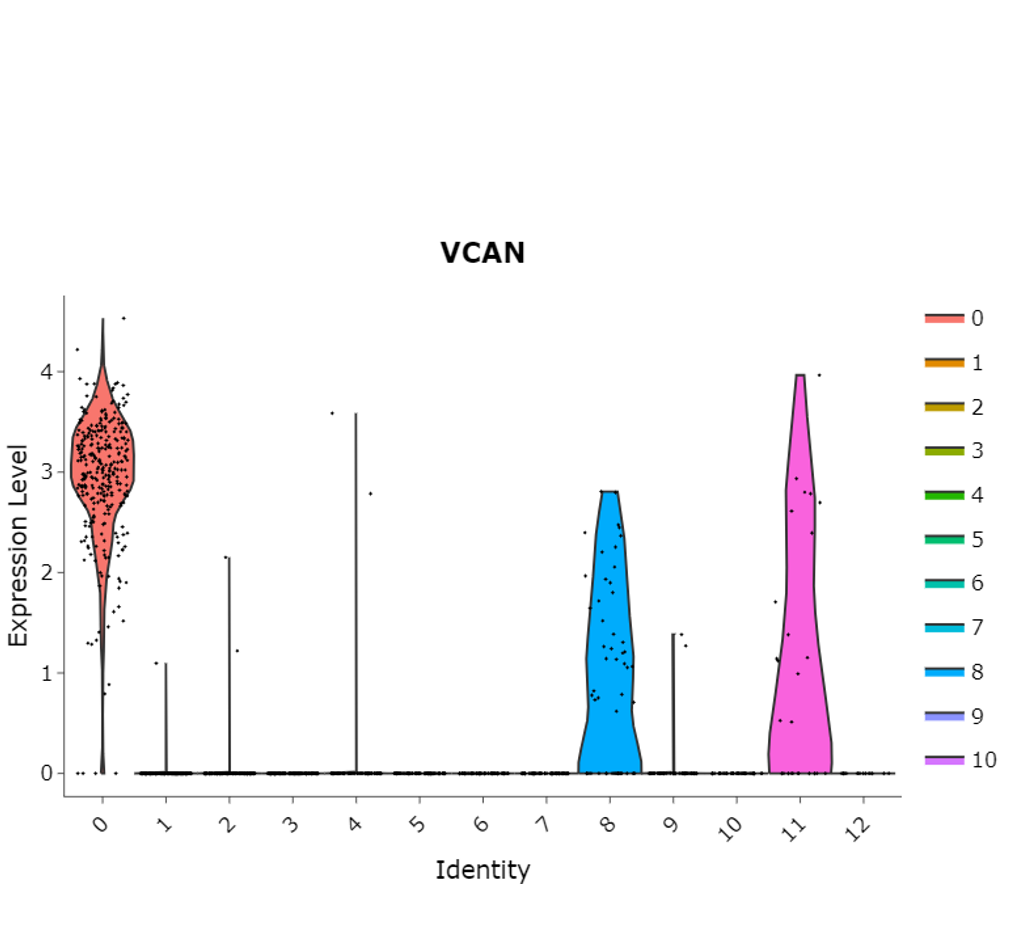
Violin Plot
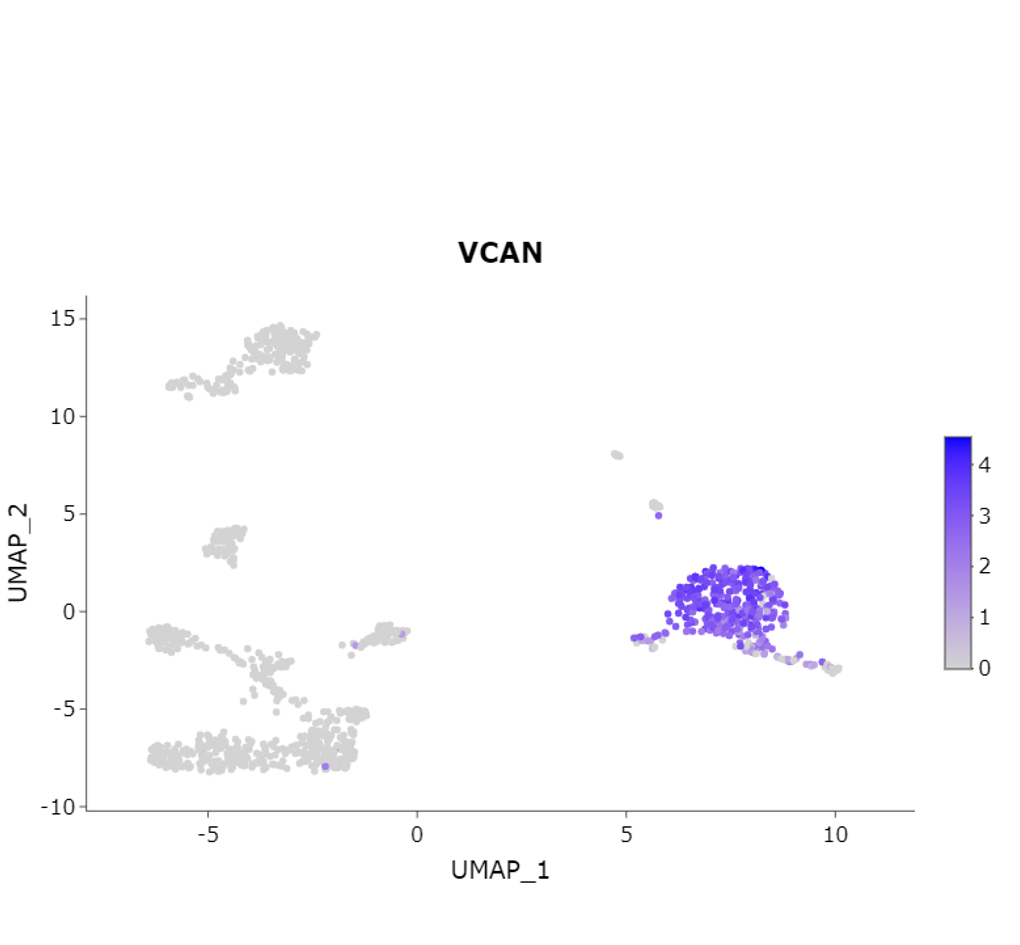
Feature Plot
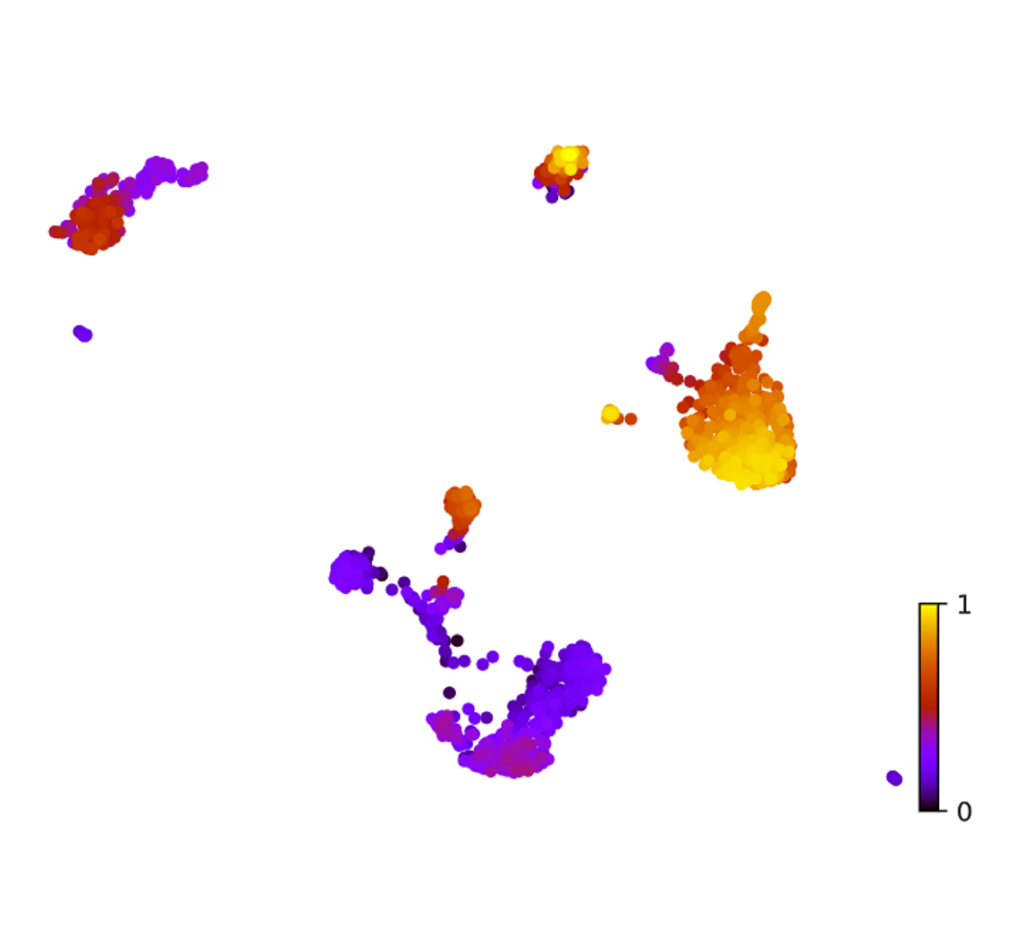
RNA Velocity
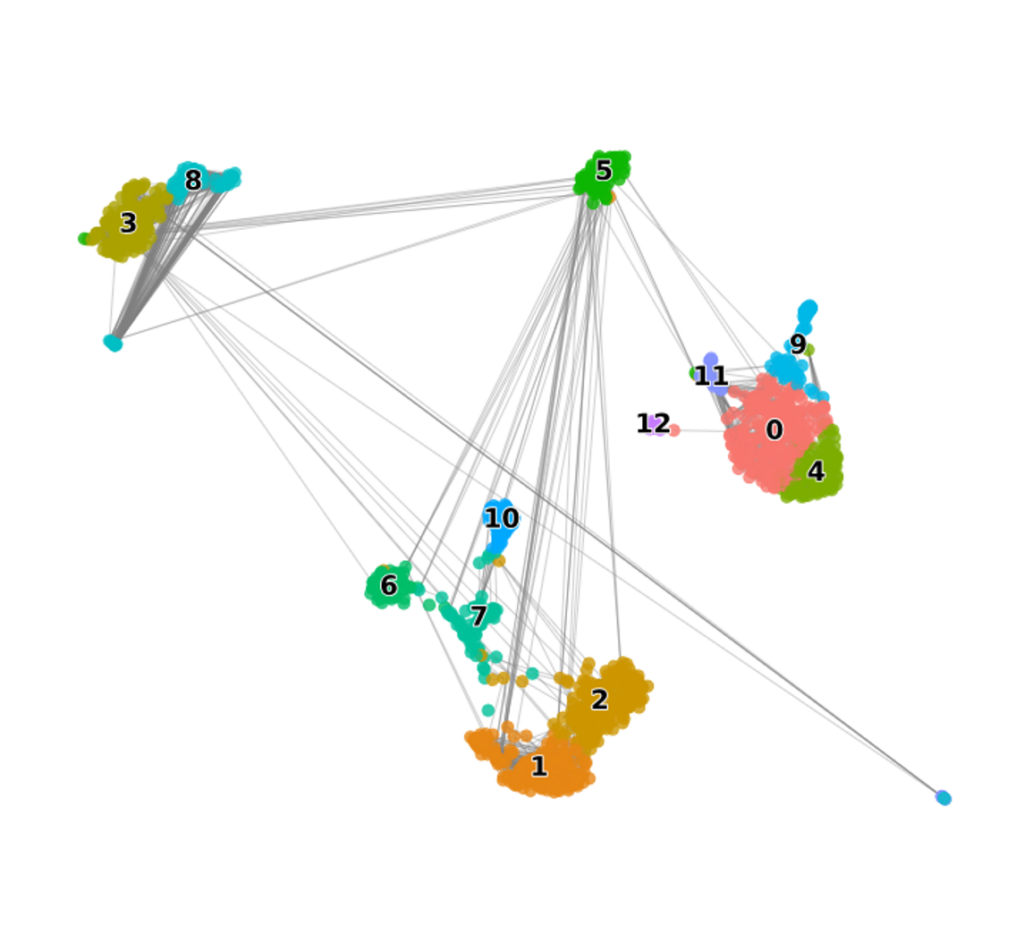
Neighbor Graph
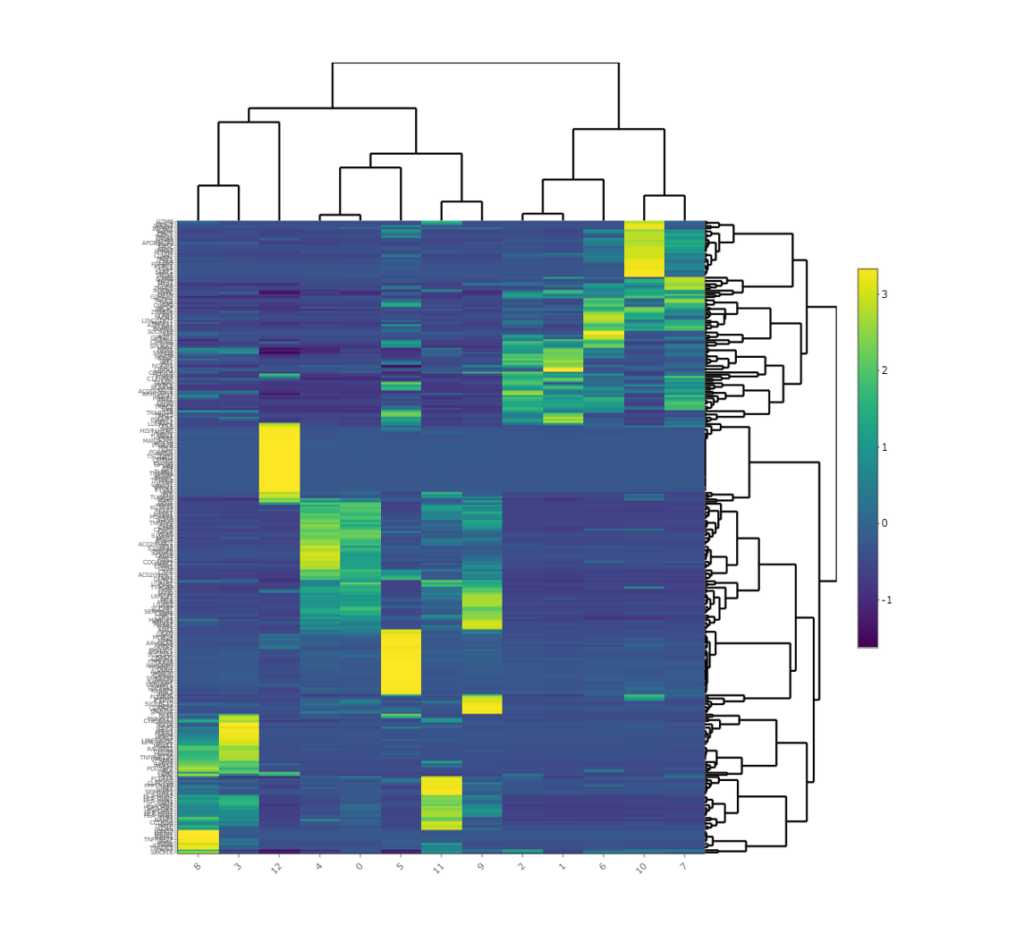
Heatmap
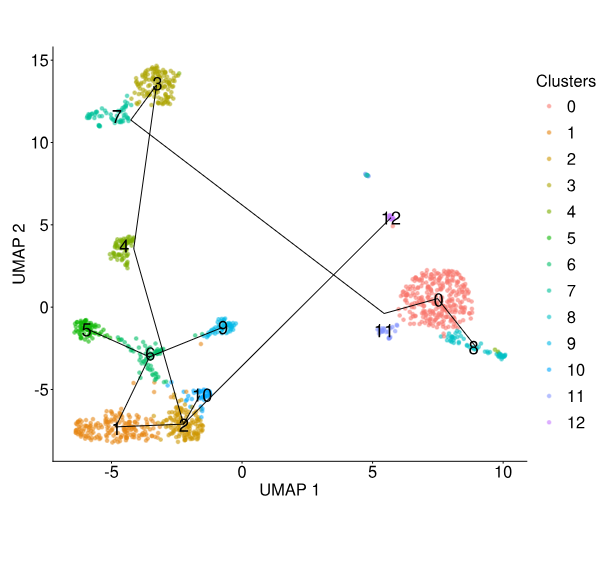
Trajectory Analysis
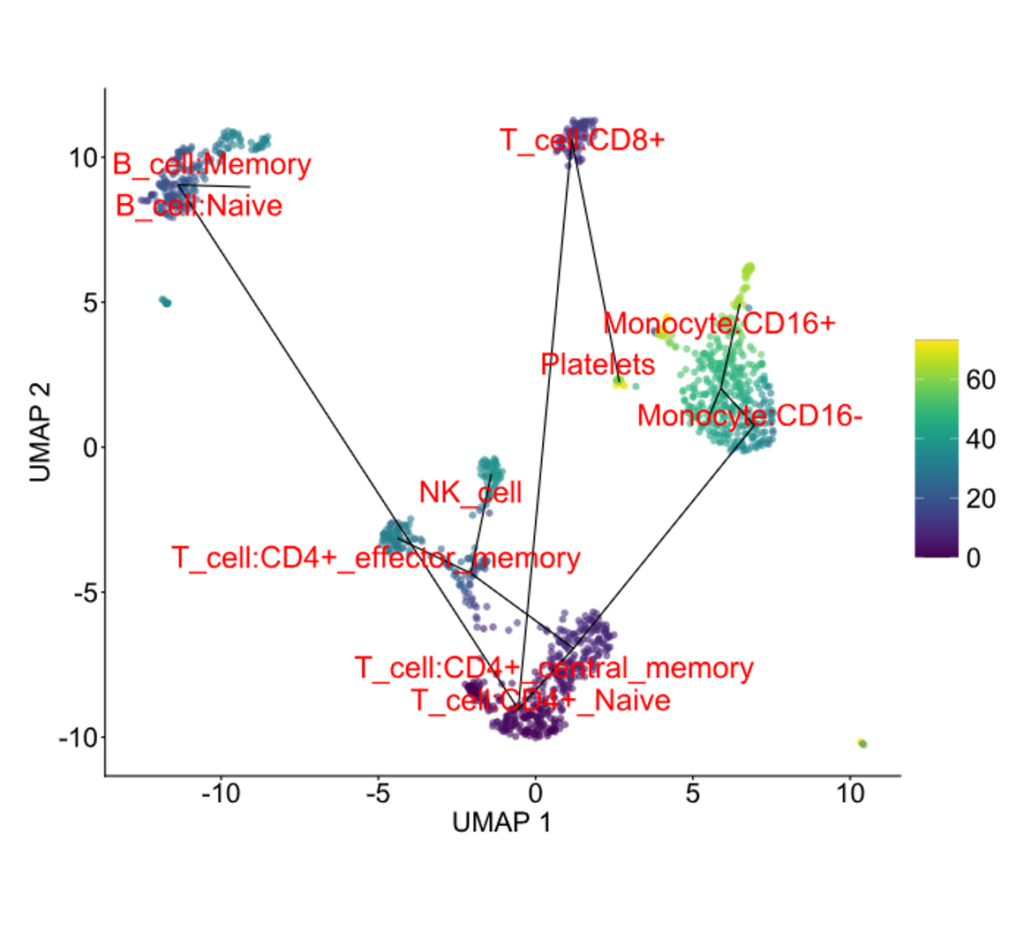
Pseudotime trajectory
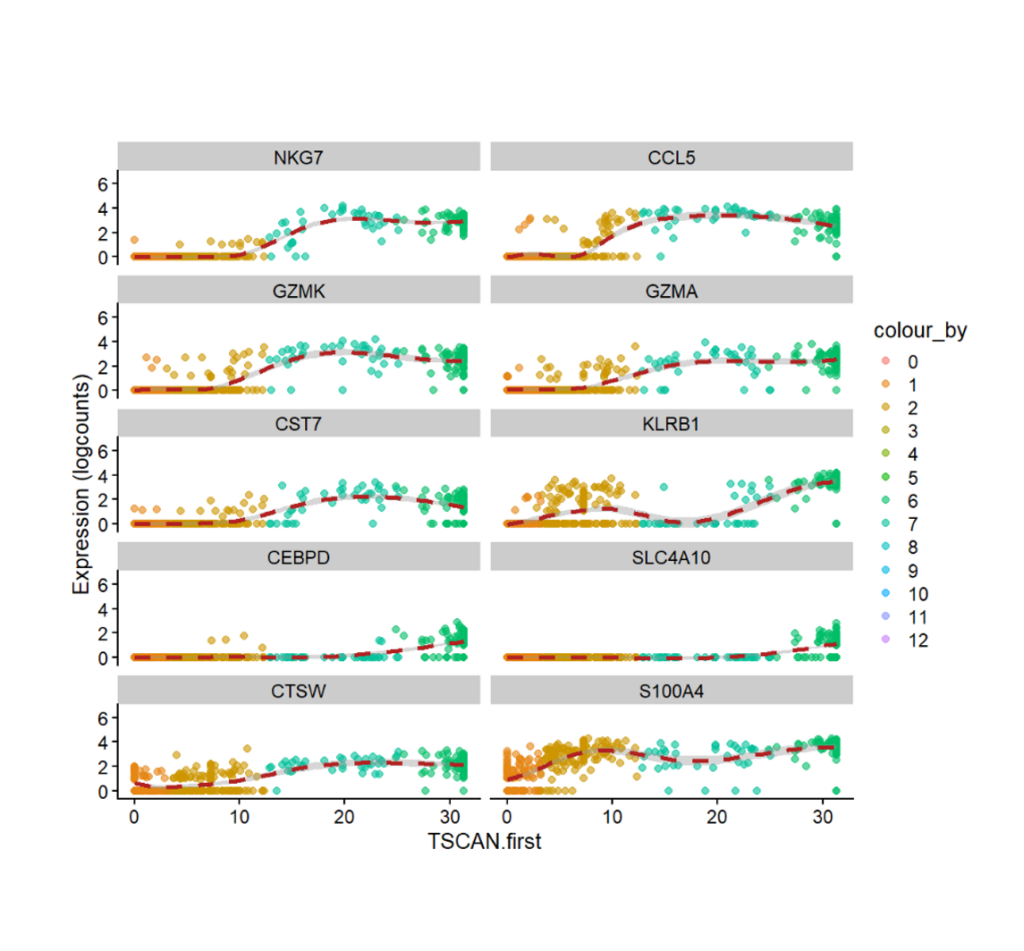
TOP 10 Expressed Genes (Up & Down) in a trajectory line
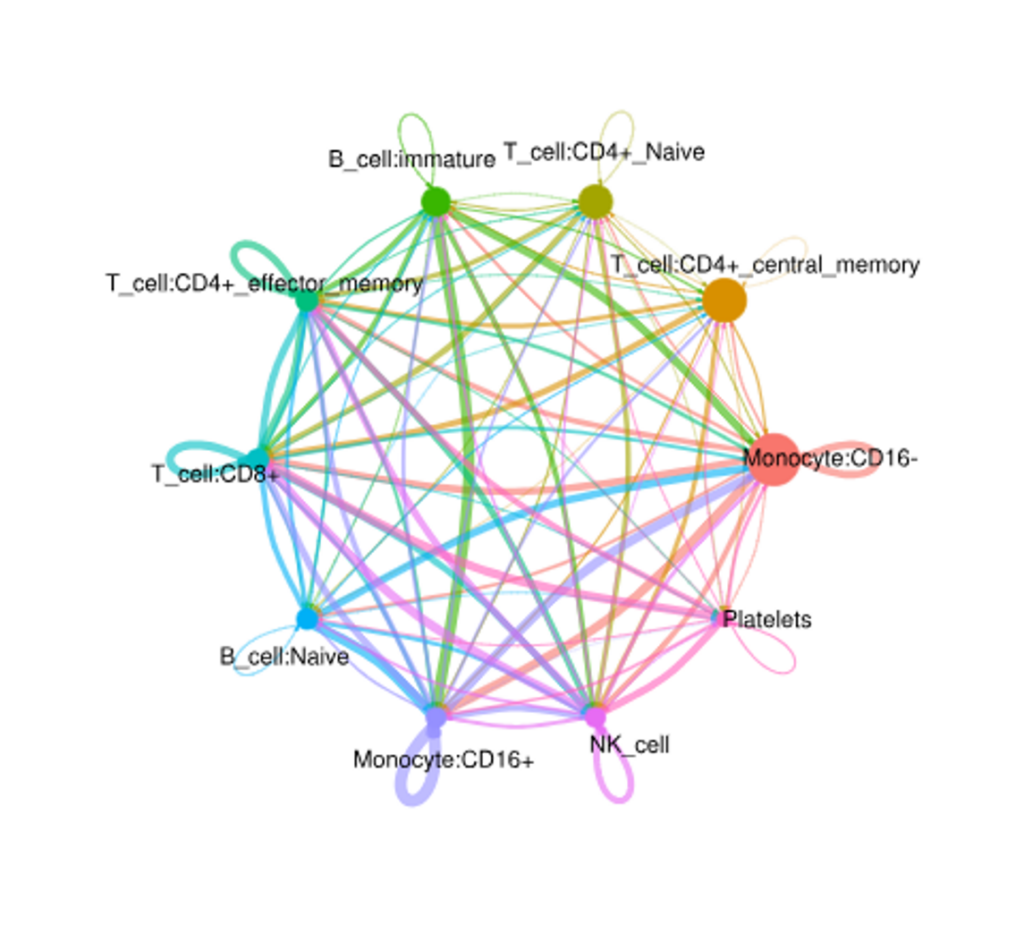
Cell chat
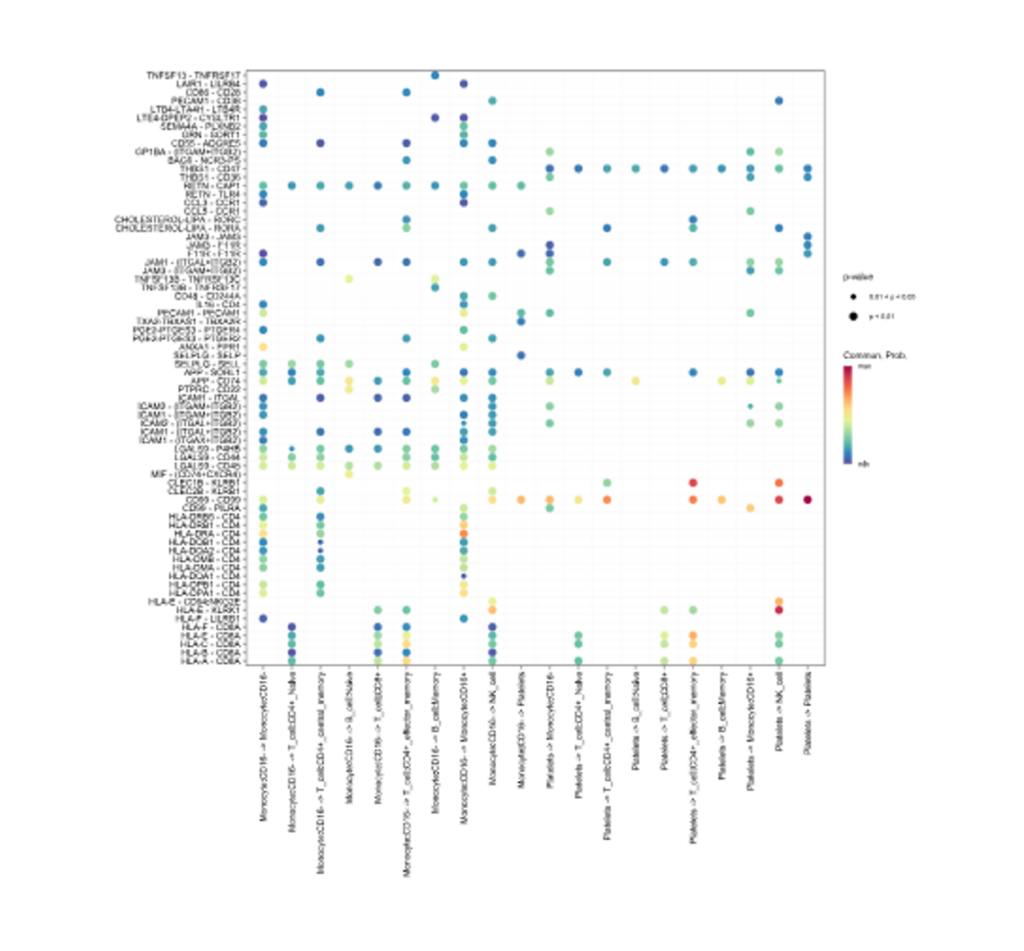
Heatmap cell chat
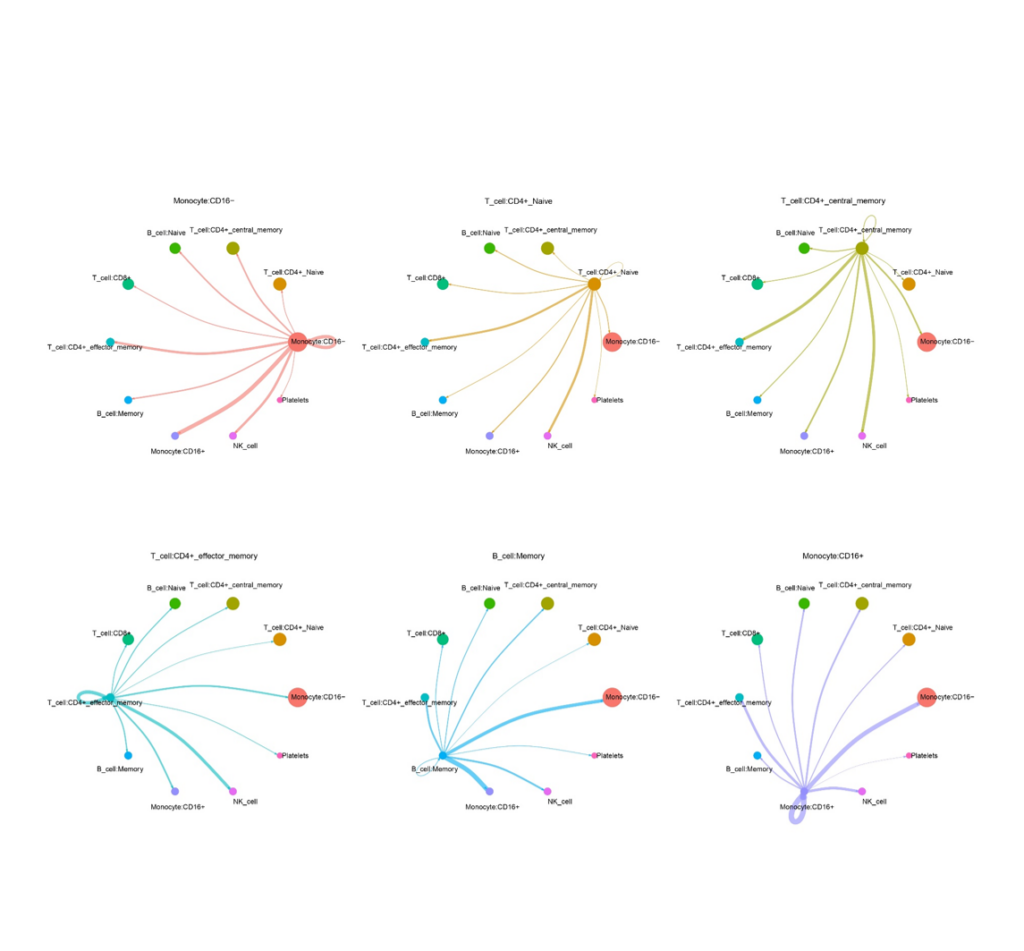
Principal Connections Between Cells
Available options
We adapt to the needs of each client through two analysis packages and the possibility of a completely customized analysis.
Standard package
- Principal Component Analysis
- Cell Clustering (2D & 3D)
- Cluster Evaluation
- Differentially Expressed Genes
- TOP Differentially Expressed Genes
- RNA Velocity
- Pathways Enrichment (GSEA & GO Terms)
- Results in Genome One Reports
Expanded package
- Everything included in the standard package
- Trajectory Analysis
- Top 10 Expressed Genes (Up & Down) in a trajectory line
- Cell Chat
- Results in Genome One Reports
"Single cell RNA-seq technology allows us to develop more precise approaches to study physiological and pathological processes at the single cell level, facilitating the identification of new biomarkers and opening new avenues of research in the diseases we work with."

Dr. Elena Sánchez Luis
Researcher

























