Transcriptomic analysis
The importance of gene expression in the study of different diseases.
Home / Transcriptomics
Study of gene expression
Transcriptomic analyses allow quantification of gene expression using NGS platforms. The interpretation of data from RNA sequencing (RNA-Seq) has become an increasingly demanded service due to its sensitivity and accuracy.
The bioinformatics analysis we perform at Dreamgenics includes:
- Quality control of the sequences.
- Alignment of reads against reference sequences.
- Quantification of gene expression in the sample.
- Differential gene expression analysis with p-value and with adjusted p-value.
- Enrichment study of gene ontologies and pathways.
- Study of isoforms generated in alternative splicing events*.
- Identification of other RNAs (smallRNAs and ncRNAs)*.
*Where the experimental design and coverage allow.

Bioinformatics Services
Results in Genome One Reports
We deliver the results of our transcriptomic analysis through our Genome One Reports platform. A simple and intuitive web platform that will provide you with the following advantages:
- You will have access to the quality analysis of the sequences obtained.
- Sample correlation analysis and differential expression study with p-value and adjusted p-value.
- PCA plot, Histogram, MA plot, Volcano plot, Heatmap and Correlation heatmap
- Enrichment of Gene Ontologies paths and terms (WikiPathways, GO-MF, GO-CC and GO-BP)
- All tables and figures are easily exportable, ready to be used in scientific publications.
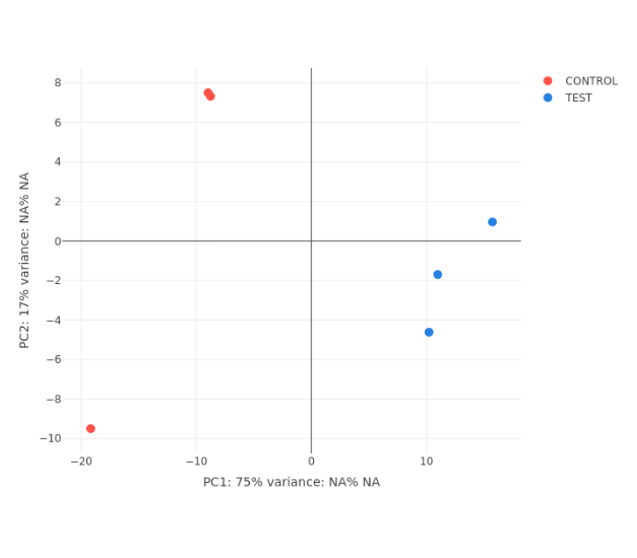
PCA Plot
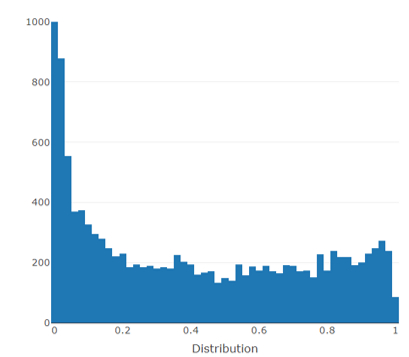
Histogram
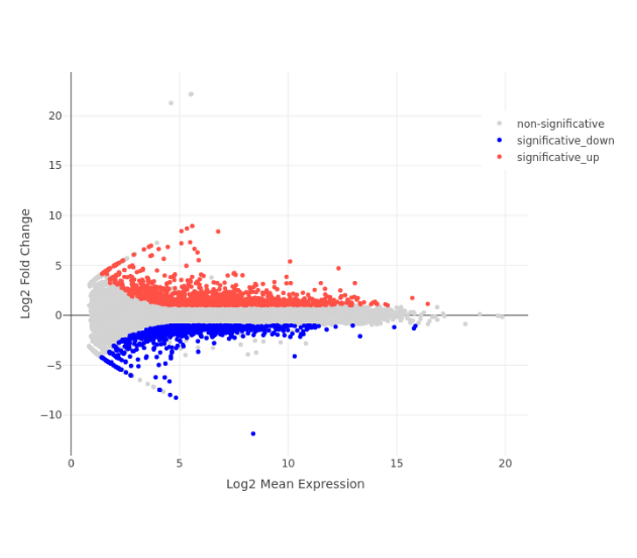
MA Plot
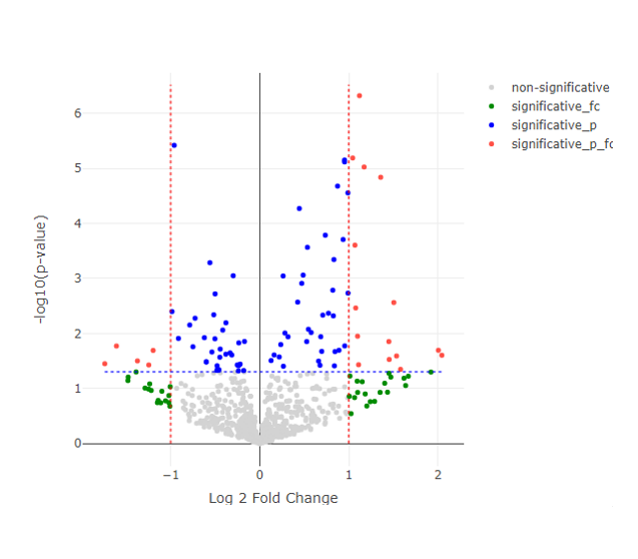
Volcano Plot
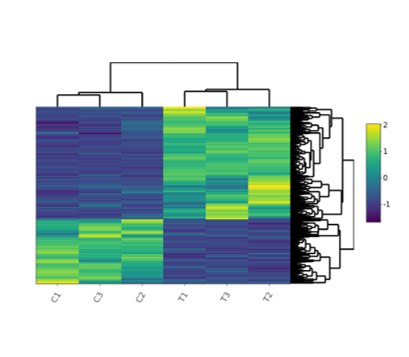
Heatmap
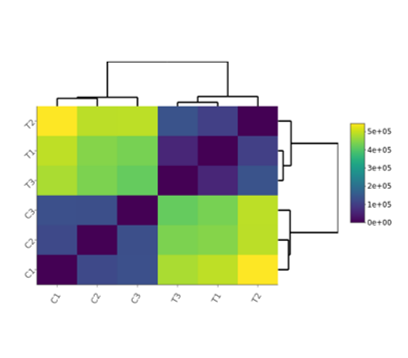
Correlation Heatmap
Different options in RNA-Seq experiments
The libraries used in an RNA-Seq experiment depend on the starting sample and the type of RNAs to be studied.
Contact us and we will help you choose the best option for your project.
mRNA-Seq & Total RNA-Seq
- DNBSEQ™-G400 Platform (IVD)
- MGIEasy Fast RNA Library
- mRNA/mRNA + lncRNA analysis
- >1ug of RNA with RIN>7
- Recommended 40-60 M/sample
- Minimum 3 samples per condition
- Differential expression, ontologies and pathways
- Results in Genome One Reports
miRNA-Seq
- DNBSEQ™-G400 Platform (IVD)
- MGIEasy Small RNA Library
- miRNA analysis
- >1ug of RNA with RIN>7
- Recommended 20 M/sample
- Minimum 3 samples per condition
- Differential expression, ontologies and pathways
- Results in Genome One Reports
*Elimination of ribosomal RNA and globin mRNA in blood samples for Total RNA-seq.


























