Genomic analysis
Relevant clinical information
Home / Genomics
Deciphering DNA information
The analysis of massive sequencing data is a complex process that requires a high level of bioinformatics knowledge and large processing and storage resources.
At Dreamgenics we have own software and pipelines for the analysis of NGS data from the sequencing of genomes, exomes, trio exomes and targeted exomes.
Our bioinformatics analysis includes:
- Quality control of the sequences.
- Alignment of reads originated against reference genome.
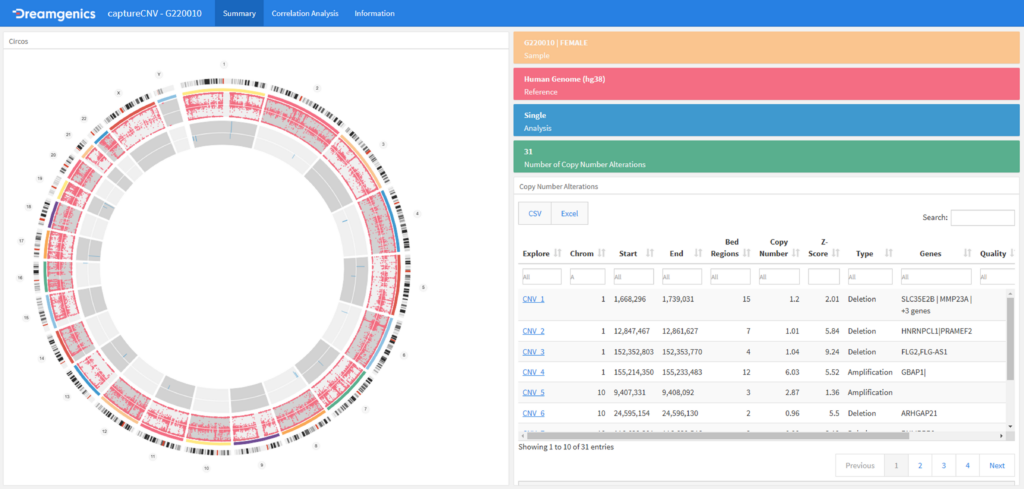
- Detection of high quality variants (SNVs, Indels and CNVs).
- Filtering of common variants in the population.
- Annotation of variants obtained with multiple databases and prediction algorithms.
- Comparison between samples and extraction of recurrent variants.

Bioinformatics Services
Results in Genome One Reports
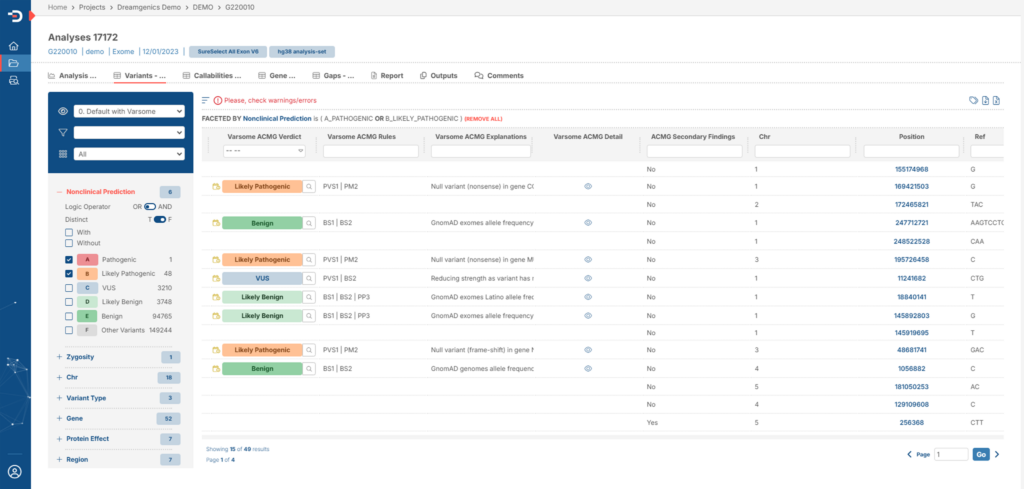
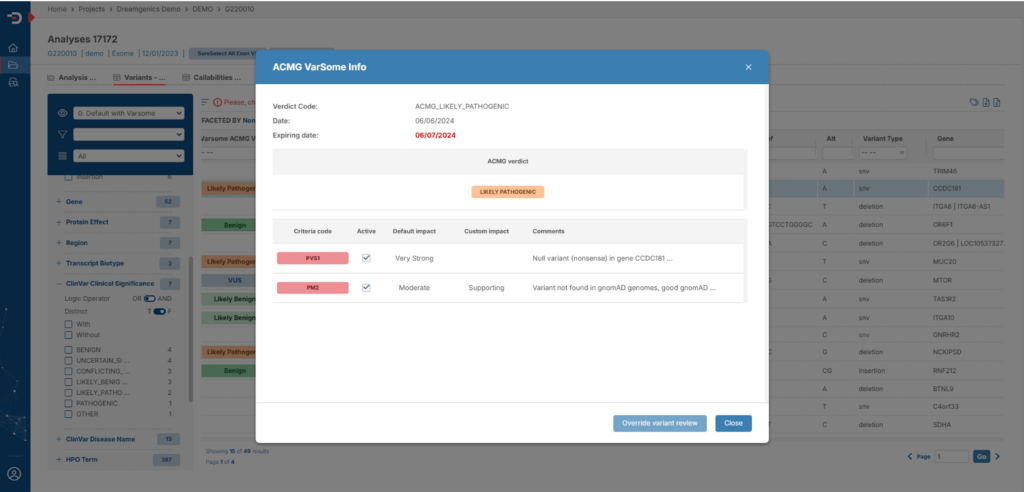
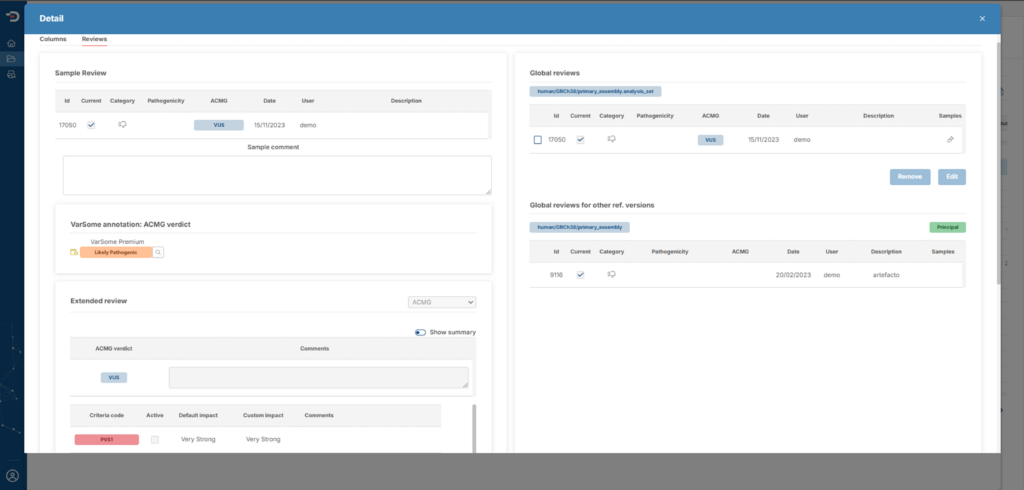
We deliver the results of our genomic analysis through our Genome One Reports platform. A simple and intuitive web platform that will provide you with the following advantages:
- You will have access to the quality analysis of the sequences obtained.
- You will be able to configure customizable variant filters
- You will have the possibility to perform a pre-sorting of variants according to pathogenicity criteria chosen by you.
- You will be able to create your own databases generated by variant review or recurrence analysis.
- You will have the option to generate customized reports
We offer a wide variety of possibilities
Depending on the starting sample and the analysis to be performed, different libraries can be used.
Contact us and we will help you choose the best option for your project.
Genome
- DNBSEQ™-G400 Platform (IVD)
- MGIEasy Fast PCR-FREE FS Library V2.0
- Mean coverage >30x
- >1 μg of genomic DNA
- 75-180 Gb/sample
- Analysis of SNVs, Indels and CNVs*.
- Results in Genome One Reports
Exome
- DNBSEQ™-G400 Platform (IVD)
- Agilent SureSelect V8/CRE V4 Library
- Mean coverage >100x
- >1 μg of genomic DNA
- 9-36 Gb/sample
- Analysis of SNVs, Indels and CNVs*.
- Results in Genome One Reports
*CNVs whenever the experimental design permits.


























